Sea urchin genetic resources
Development of Lytechinus pictus
The sea urchin embryo has a rich history as a developmental model system. One of the most frequently used species is the purple sea urchin, Strongylocentrotus purpuratus. However, the long generation time (2 years) for this organism has prevented researchers from maximizing the potential of the urchin embryo model. We are developing the painted white urchin, Lytechinus pictus, as an alternative urchin embryo system. The shorter generation time (6 months) and faster pace of development of this species allows greater access to later life history stages, and enables more powerful manipulations of this system over time.
Molecular Advances
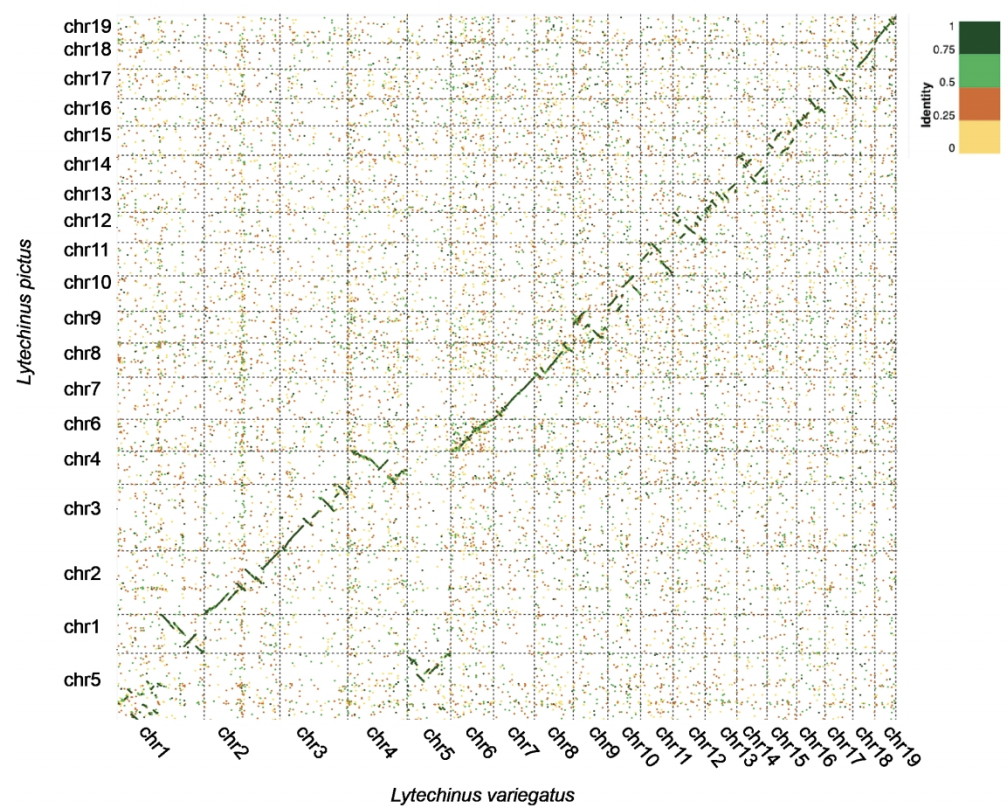
To realize the potential of a new urchin model species, we have generated a transcriptome from pooled developmental stages, and are in progress of annotating the L. pictus genome in collaboration with the Lyons lab among others. The genome will be hosted online and publicly available through Echinobase shortly. This works is in collaboration with the Lyons Lab at SIO.
Transgenesis
Transgenic organisms have contributed greatly to biological research, and serve as powerful tools. We are harnessing the power of CRISPR genome editing technology to modify the genome of the sea urchin. We are in progress of generating inbred lines of Lytechinus pictus with stable genetic backgrounds to further enhance the efficiency of genomic manipulations and the utility of L. pictus as a model species of sea urchin. A custom marine transgenics facility in Hubbs Hall at SIO will facilitate maintaining these lines. Our labs have recently described updated culturing methods for this species.
Gene-editing and knock-ins
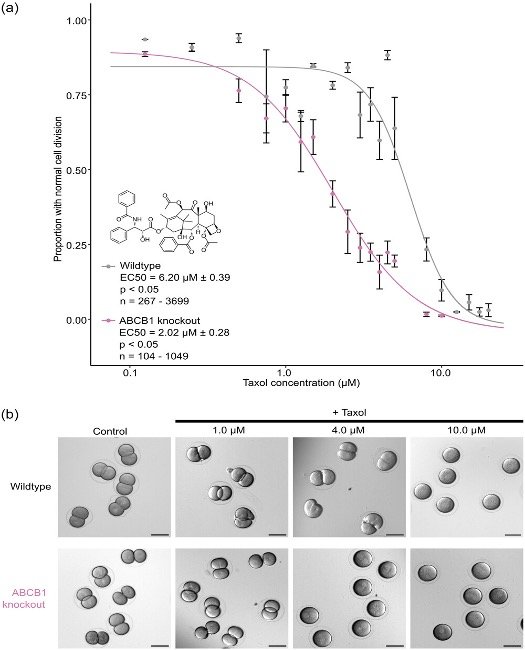
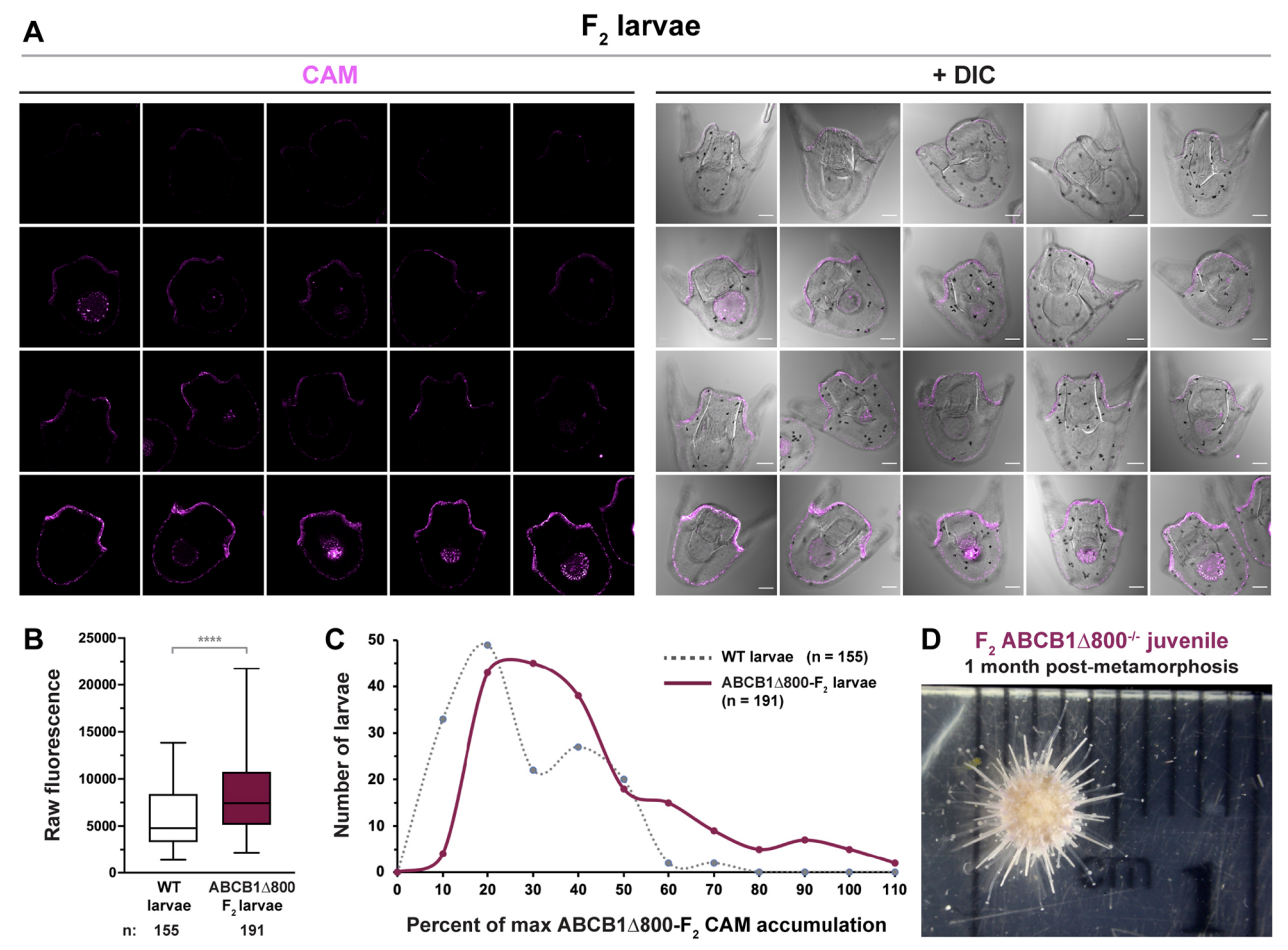
The Hamdoun lab utilizes CRISPR/Cas9 genome editing technology for creating loss-of-function mutations in xenobiotic transporters to understand their potential roles in early sea urchin development. These gene knockouts will also help us understand how sea urchins protect themselves against exposures to anthropogenic chemicals in the marine habitat. We are also using CRISPR and transposon mutagenesis to insert fluorescent protein markers into the L. pictus and S. purpuratus genomes. These techniques will allow us to understand transcriptional regulation, label and manipulate endogenous proteins, and isolate cell-types of interest. As we expand this program it will become a resource for researchers interested in development, xenobiotic stress, and more.